5.3. Direct Linear Transform (DLT)#
This notebook demonstrates the Direct Linear Transform (DLT) method for camera calibration, following standard computer vision notation. The DLT provides a linear solution to estimate the camera projection matrix P from known 3D-2D point correspondences.
Learning Objectives: You’ll learn how to estimate a camera’s projection matrix from known 3D-2D point correspondences using the Direct Linear Transform (DLT) algorithm, understand camera matrix decomposition into intrinsic and extrinsic parameters and analyze calibration quality through reprojection error metrics.
5.3.1. Prerequisites#
Note
Before reading this chapter you should be comfortable with matrices, vectors, homogeneous transformations, and basic concepts of linear algebra. See chapter Algebra & Calculus for a primer on these foundations.
5.3.2. Camera Projection Model#
5.3.2.1. Pinhole Camera Geometry#
The pinhole camera model relates 3D world coordinates to 2D image coordinates through perspective projection. Please consult Pinhole Camera Model for detailed information.
We define the homogeneous coordinates for 3D world point \(X\) and image point \(x\):
now we can write the relation using the projection matrix \(P\)
or in proportional (hiding the scale ambiguity) description
where:
\((u,v)\) are pixel coordinates in the image
\((X,Y,Z)\) are 3D world coordinates
\(s\) is a scale factor (projective depth)
\(\mathbf{P}\) is the \(3 \times 4\) projection matrix
5.3.2.2. Projection Matrix Decomposition#
The projection matrix can be decomposed as:
where:
\(\mathbf{K} \in \mathbb{R}^{3 \times 3}\) is the intrinsic matrix: \(\mathbf{K} = \begin{bmatrix} f_x & \alpha & c_x \\ 0 & f_y & c_y \\ 0 & 0 & 1 \end{bmatrix}\)
\(f_x, f_y\): focal lengths in pixels
\(c_x, c_y\): principal point coordinates
\(\alpha\): skew parameter (usually ≈ 0)
\(\mathbf{R} \in \mathbb{R}^{3 \times 3}\) is the rotation matrix (world to camera)
\(\mathbf{t} \in \mathbb{R}^{3 \times 1}\) is the translation vector (camera position in world coordinates)
\([\mathbf{R}|\mathbf{t}]\) represents the extrinsic parameters (camera pose)
The projection equation becomes:
5.3.2.3. Degrees of Freedom#
The projection matrix \(\mathbf{P}\) has:
12 entries total
1 scale ambiguity (homogeneous representation)
11 degrees of freedom
Therefore, we need ≥ 6 point correspondences (each provides 2 constraints) to solve for \(\mathbf{P}\) up to scale \(s\).
5.3.2.4. Generate Synthetic 3D World Points and Camera Parameters#
We create:
Random 3D points in a cube
Intrinsic matrix \(\mathbf{K}\) with \(f_x\), \(f_y\), \(\alpha\) (set 0), principal point (\(c_x\), \(c_y\))
Rotation from Euler angles (\(\mathbf{R_x}\), \(\mathbf{R_y}\), \(\mathbf{R_z}\))
Translation vector \(t\)
Camera matrix \(\mathbf{P} = \mathbf{K}[\mathbf{R}|\mathbf{t}]\)
All points are represented in homogeneous coordinates when needed.
def euler_xyz(rx, ry, rz):
"""Compose rotation matrix from Euler angles (radians)."""
Rx = np.array(
[[1, 0, 0], [0, np.cos(rx), -np.sin(rx)], [0, np.sin(rx), np.cos(rx)]]
)
Ry = np.array(
[[np.cos(ry), 0, np.sin(ry)], [0, 1, 0], [-np.sin(ry), 0, np.cos(ry)]]
)
Rz = np.array(
[[np.cos(rz), -np.sin(rz), 0], [np.sin(rz), np.cos(rz), 0], [0, 0, 1]]
)
return Rz @ Ry @ Rx # Z-Y-X order
def generate_intrinsics(fx=1200, fy=1180, cx=640, cy=360, alpha=0.0):
K = np.array([[fx, alpha, cx], [0.0, fy, cy], [0.0, 0.0, 1.0]])
return K
5.3.2.5. Exercies 1: Create a function to create the projection_matrix#
Fill the given template to construct the projection matrix
Test it in the following cell
def assemble_projection_matrix(K, R, t):
"""Return 3x4 camera matrix P = K [R|t]."""
pass
# Function to generate random 3D scene points
def generate_scene(n_points=40, seed=42, cube_size=2.0):
rng = np.random.default_rng(seed)
Xw = rng.uniform(-cube_size, cube_size, (n_points, 3))
return Xw
# Example camera
rx, ry, rz = np.deg2rad([10, -5, 20])
R_gt = euler_xyz(rx, ry, rz)
t_gt = np.array([0.5, -0.2, 6.0]) # camera looking roughly toward origin
K_gt = generate_intrinsics()
P_gt = assemble_projection_matrix(K_gt, R_gt, t_gt)
X_world = generate_scene(n_points=6, seed=42)
print("Ground truth camera matrix P:\n", P_gt)
print()
print("First 2 world points:\n", X_world[:2])
Ground truth camera matrix P:
[[ 1.17911984e+03 -3.10543036e+02 6.02361534e+02 4.44000000e+03]
[ 4.33424078e+02 1.14815901e+03 1.25993858e+02 1.92400000e+03]
[ 8.71557427e-02 1.72987394e-01 9.81060262e-01 6.00000000e+00]]
First 2 world points:
[[ 1.09582419 -0.24448624 1.43439168]
[ 0.78947212 -1.62329061 1.90248941]]
5.3.2.6. Exercies 2: Create a function to project points#
Write a function that calculates the 3D projections.
It requires the projection matrix \(\mathbf{P}\) and set of 3D points \(X\) as input arguments.
Run it on the generated 3D world points.
def project_points(P, X):
"""Project (N,3) world points X using camera P (3x4). Return (N,2) image points."""
pass
x_gt, xh_gt = project_points(P_gt, X_world)
print("Sample projected points of the first 2 world points:\n", x_gt[:2])
Sample projected points of the first 2 world points:
[[894.32457186 308.15506282]
[917.23973743 83.88349871]]
5.3.3. 4. DLT Algorithm: Linear Solution#
The Direct Linear Transform (DLT) method exploits the fundamental property that for any 3D point \(X\) projecting to image point \(x\) equation (5.31) is valid. This means the vectors \(\tilde{x}\) and \(P \tilde{X}\) are parallel (or anti-parallel).
5.3.3.1. Cross Product Property#
Note
For a primer on vector cross products, see our Vector Cross Product chapter.
Since parallel vectors have zero cross product:
This is the key insight that transforms the projective equation into a linear homogeneous system.
Let’s expand (5.34) systematically:
The projection gives:
Now equation (5.34) can be written as:
5.3.3.2. Rearranging into Standard DLT Form#
Rearranging the first two lines of (5.35) yields two equation. The third one is irrelevant as it is a linear combination of the first two.
Equation 1:
Equation 2:
We want the rewrite the previous equation as a linear system of equations in the standard form of \(\mathrm{A} p = b\), with \(b=0\) and \(p = [p_1^T, p_2^T, p_3^T]^T\) as a 12-element vector, each equation becomes a row in matrix \(A\):
Row 1 (from equation 2):
Row 2 (from equation 1):
Essentially we have 2 equations per measurement point and 12 unknowns of the \(\mathrm{P}\) matrix.
The DLT method essentially asks: “What camera matrix makes all observed image rays parallel to their corresponding projected 3D points?”
This cross product formulation is the mathematical foundation that makes camera calibration a linear algebra problem solvable via SVD!
5.3.3.3. Exercise 3: Create a function to create the DLT (or A) matrix#
Use the 3D points and 2D image points as inputs
For each measurement create 2 rows
def build_dlt_matrix(X, x):
"""Build DLT design matrix A from 3D points X (N,3) and image points x (N,2)."""
N = X.shape[0]
A = []
for i in range(N):
pass
A = build_dlt_matrix(X_world, x_gt)
print("A shape:", A.shape)
print("First 2 rows:\n", A[:2])
A shape: (12, 12)
First 2 rows:
[[ 1.09582419e+00 -2.44486241e-01 1.43439168e+00 1.00000000e+00
0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
-9.80022503e+02 2.18650053e+02 -1.28281172e+03 -8.94324572e+02]
[ 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
1.09582419e+00 -2.44486241e-01 1.43439168e+00 1.00000000e+00
-3.37683773e+02 7.53396730e+01 -4.42015058e+02 -3.08155063e+02]]
5.3.3.4. Solve Homogeneous System via SVD#
Read the information on Column Space and Null Space to gain information on null spaces. The null space is the solution to the aforementioned equation \(\mathrm{A} p = 0\).
We compute the singular value decomposition (SVD):
The null space vector p, i.e. the solution of matrix \(A\) (up to scale), is the last column of V corresponding to the smallest singular value.
Connects to earlier rank/null space concepts:
Expect \(\mathrm{rank}(A) = 11\) (nullity 1) when there are sufficient non-degenerate points (≥6).
def solve_dlt(A, analyze=True):
"""Solve A p = 0 via SVD, return p (length 12)."""
U, S, Vt = svd(A)
p = Vt[-1] # last row of V^T, i.e., last column of V
if analyze:
rankA = np.sum(S > 1e-10)
cond = (
S[0] / S[-2]
) # ignore last singular value which is (near) zero in our dlt case
print("Design matrix shape:", A.shape)
print()
print("Rank(A):", rankA, "Nullity:", A.shape[1] - rankA)
print()
print("Singular values:\n", S)
print()
print("Condition number:", cond)
print()
res = norm(A @ p)
print("||A p||_2:", res)
return p
p_est_raw = solve_dlt(A, analyze=True)
Design matrix shape: (12, 12)
Rank(A): 11 Nullity: 1
Singular values:
[3.85223749e+03 2.84321198e+03 1.14471066e+03 9.91327067e+02
4.21212466e+00 2.80364060e+00 1.84955587e+00 1.20674556e+00
6.13219176e-01 3.03575643e-01 2.69256383e-01 3.84821912e-15]
Condition number: 14306.949527170735
||A p||_2: 1.355399879558074e-13
5.3.3.5. Reshape Parameter Vector into Camera Matrix#
Reshape p into P_est (3×4). Because the homogeneous system yields scale ambiguity, enforce a scale convention (e.g. \(||P_{est}[2,:]|| = 1\) or divide by last element if sensible).
def vector_to_camera(p):
P = p.reshape(3, 4)
# Scale convention: normalize third row norm
scale = norm(P[2, :])
P /= scale
return P
P_est = vector_to_camera(p_est_raw)
print("Estimated camera matrix P_est:\n", P_est)
print("Ground truth (scaled for comparison):\n", P_gt / norm(P_gt[2, :]))
Estimated camera matrix P_est:
[[-1.93846108e+02 5.10529606e+01 -9.90276262e+01 -7.29931504e+02]
[-7.12544796e+01 -1.88756179e+02 -2.07132626e+01 -3.16303652e+02]
[-1.43283158e-02 -2.84389524e-02 -1.61285314e-01 -9.86393924e-01]]
Ground truth (scaled for comparison):
[[ 1.93846108e+02 -5.10529606e+01 9.90276262e+01 7.29931504e+02]
[ 7.12544796e+01 1.88756179e+02 2.07132626e+01 3.16303652e+02]
[ 1.43283158e-02 2.84389524e-02 1.61285314e-01 9.86393924e-01]]
5.3.4. 7. Camera Matrix Decomposition#
5.3.4.1. RQ Factorization#
The estimated camera matrix can be decomposed in a orthonormal part and upper triangular part which correspond with the extrinsic and intrinsic part. Decomposing can be done using the QR or RQ decomposition methods.
Given \(\mathbf{P} = [\mathbf{M} | \mathbf{m}_4]\) where \(\mathbf{M} = \mathbf{K}\mathbf{R}\) is \(3 \times 3\):
where:
\(\mathbf{K}\) is upper triangular (intrinsic parameters)
\(\mathbf{R}\) is orthogonal (rotation matrix)
5.3.4.2. Intrinsic Matrix Structure#
\(f_x, f_y\): focal lengths (pixels)
\(c_x, c_y\): principal point coordinates
\(\alpha\): skew parameter (usually ≈ 0)
5.3.4.3. Translation Recovery#
Once \(\mathbf{K}\) and \(\mathbf{R}\) are known: $\(\mathbf{t} = \mathbf{K}^{-1} \mathbf{m}_4\)$
Note: Translation is recovered up to scale (fundamental limitation of projective geometry).
def decompose_camera_matrix(P):
"""Decompose camera matrix into K, R, t using RQ on left 3x3."""
M = P[:, :3]
K, R = sla.rq(M) # R is orthogonal, K is upper-triangular
# Enforce positive diagonal
T = np.diag(np.sign(np.diag(K)))
K = K @ T
R = T @ R
# Normalize K (K[2,2] should be 1)
K /= K[2, 2]
t = np.linalg.inv(K) @ P[:, 3]
return K, R, t
K_est, R_est, t_est = decompose_camera_matrix(P_est)
print("Estimated K:\n", K_est)
print("Ground truth K:\n", K_gt)
print("\nRotation error (Fro norm):", norm(R_est - R_gt, ord="fro"))
print("Translation est:", t_est)
print("Translation gt:", t_gt)
Estimated K:
[[1.20000000e+03 3.40178182e-10 6.40000000e+02]
[0.00000000e+00 1.18000000e+03 3.60000000e+02]
[0.00000000e+00 0.00000000e+00 1.00000000e+00]]
Ground truth K:
[[1.20e+03 0.00e+00 6.40e+02]
[0.00e+00 1.18e+03 3.60e+02]
[0.00e+00 0.00e+00 1.00e+00]]
Rotation error (Fro norm): 3.4641016151377553
Translation est: [-0.08219949 0.0328798 -0.98639392]
Translation gt: [ 0.5 -0.2 6. ]
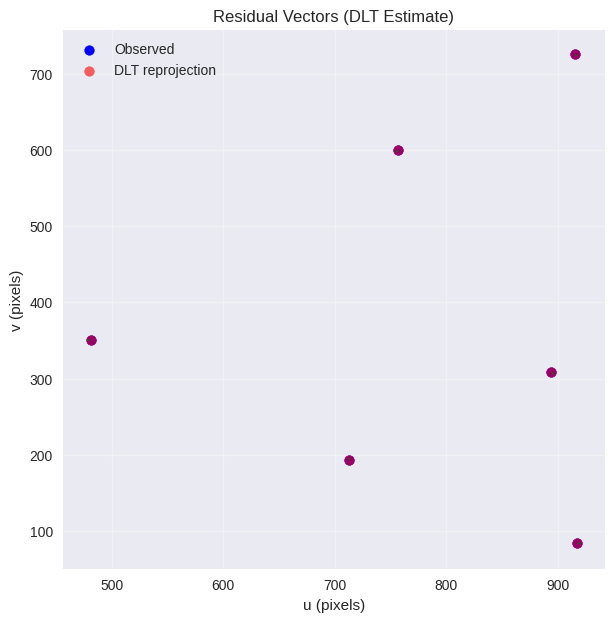
5.3.5. Reprojection Error Evaluation#
The reprojection error measures how well our estimated camera matrix P reproduces the observed 2D points from known 3D points.
Definition: For each 3D-2D correspondence \((X_i, x_i)\):
Project 3D point: \(\hat{x}_i = \text{project}(P, X_i)\)
Compute Euclidean distance: \(e_i = \|x_i - \hat{x}_i\|_2\)
Geometric Meaning: Reprojection error represents the pixel distance between:
Observed image points \(x_i\) (ground truth)
Predicted image points \(\hat{x}_i\) (from estimated camera)
Quality Metrics:
Mean error: Average distance across all points
RMS error: \(\sqrt{\frac{1}{n}\sum e_i^2}\) (standard calibration metric)
Max error: Worst-case correspondence (outlier detection)
We compute mean, RMS, max errors and visualize residual vectors.
def reprojection(P, X):
x, _ = project_points(P, X)
return x
def reprojection_error(P, X, x_obs):
x_est = reprojection(P, X)
residuals = x_est - x_obs
errs = np.sqrt(np.sum(residuals**2, axis=1))
stats = {
"mean": np.mean(errs),
"rms": np.sqrt(np.mean(errs**2)),
"max": np.max(errs),
}
return stats, residuals
stats_gt, _ = reprojection_error(P_gt, X_world, x_gt)
stats_est, residuals_est = reprojection_error(P_est, X_world, x_gt)
print("Ground truth reprojection stats (should be near zero):", stats_gt)
print("DLT reprojection stats:", stats_est)
# Plot residuals
plt.figure(figsize=(7, 7))
plt.scatter(x_gt[:, 0], x_gt[:, 1], c="blue", label="Observed")
x_dlt = reprojection(P_est, X_world)
plt.scatter(x_dlt[:, 0], x_dlt[:, 1], c="red", alpha=0.6, label="DLT reprojection")
for i in range(len(x_gt)):
plt.arrow(
x_gt[i, 0],
x_gt[i, 1],
residuals_est[i, 0],
residuals_est[i, 1],
color="green",
head_width=2.0,
length_includes_head=True,
alpha=0.5,
)
plt.title("Residual Vectors (DLT Estimate)")
plt.xlabel("u (pixels)")
plt.ylabel("v (pixels)")
plt.legend()
plt.grid(alpha=0.3)
plt.show()
Ground truth reprojection stats (should be near zero): {'mean': 0.0, 'rms': 0.0, 'max': 0.0}
DLT reprojection stats: {'mean': 3.89559070460572e-11, 'rms': 4.462257296118922e-11, 'max': 6.488033381345533e-11}
5.3.6. Rank, Null Space, and Conditioning Analysis#
We analyze matrix A:
Rank should be 11 for sufficient non-degenerate points
Singular spectrum gives conditioning insight
Null space dimension 1 mirrors earlier column-space-null-space discussions
Large condition numbers indicate sensitivity to noise.
solve_dlt(A, analyze=True)
# solve_dlt(A_norm)
X_world_ = generate_scene(n_points=200, seed=4)
x_gt_, _ = project_points(P_gt, X_world_)
solve_dlt(build_dlt_matrix(X_world_, x_gt_), analyze=True)
Design matrix shape: (12, 12)
Rank(A): 11 Nullity: 1
Singular values:
[3.85223749e+03 2.84321198e+03 1.14471066e+03 9.91327067e+02
4.21212466e+00 2.80364060e+00 1.84955587e+00 1.20674556e+00
6.13219176e-01 3.03575643e-01 2.69256383e-01 3.84821912e-15]
Condition number: 14306.949527170735
||A p||_2: 1.355399879558074e-13
Design matrix shape: (400, 12)
Rank(A): 11 Nullity: 1
Singular values:
[1.73520571e+04 1.50940812e+04 1.40818549e+04 9.29858004e+03
1.82332070e+01 1.64449042e+01 1.51086987e+01 1.35289715e+01
6.44711320e+00 6.11522285e+00 5.16813784e+00 2.69580147e-15]
Condition number: 3357.5066397070864
||A p||_2: 2.1188397530174233e-12
array([-2.27822075e-01, 6.00011605e-02, -1.16384484e-01, -8.57868707e-01,
-8.37434580e-02, -2.21840019e-01, -2.43437360e-02, -3.71743106e-01,
-1.68396811e-05, -3.34235297e-05, -1.89554256e-04, -1.15928204e-03])
5.3.7. Degeneracy and Minimal Configuration Tests#
DLT for a 3x4 camera requires at least 6 point correspondences (12 equations) but in practice more for stability.
Degenerate cases:
All points coplanar (world points in a plane) reduce information
Points arranged on a line
Insufficient number of points
We demonstrate rank drop and inflated condition numbers.
def degeneracy_demo():
def make_coplanar_points(n=20, z=0.0, size=4.0):
rng = np.random.default_rng(1)
X = rng.uniform(-size, size, (n, 2))
X3 = np.hstack([X, np.full((n, 1), z)])
return X3
X_plane = make_coplanar_points()
x_plane, _ = project_points(P_gt, X_plane)
A_plane = build_dlt_matrix(X_plane, x_plane)
print("Coplanar case:")
solve_dlt(A_plane)
X_line = np.array([[i * 0.1, 0, 0] for i in range(20)])
x_line, _ = project_points(P_gt, X_line)
A_line = build_dlt_matrix(X_line, x_line)
print("\nCollinear case:")
solve_dlt(A_line)
X_few = X_world[:5]
x_few, _ = project_points(P_gt, X_few)
A_few = build_dlt_matrix(X_few, x_few)
print("\nToo few points (5 world points -> 10 rows):")
solve_dlt(A_few)
def ill_conditioned_demo():
print("\n3D very clos:e")
X_world = generate_scene(n_points=40, seed=4, cube_size=0.01)
x_gt, _ = project_points(P_gt, X_world)
solve_dlt(build_dlt_matrix(X_world, x_gt), analyze=True)
print("\n3D very far apart")
X_world = generate_scene(n_points=40, seed=4, cube_size=1000.0)
x_gt, _ = project_points(P_gt, X_world)
solve_dlt(build_dlt_matrix(X_world, x_gt), analyze=True)
print("\n--- Degeneracy Demo ---")
degeneracy_demo()
print("\n--- Ill-Conditioned Demo ---")
ill_conditioned_demo()
--- Degeneracy Demo ---
Coplanar case:
Design matrix shape: (40, 12)
Rank(A): 8 Nullity: 4
Singular values:
[1.14838991e+04 8.69010371e+03 3.69218495e+03 9.85399430e+00
8.86537882e+00 7.62252352e+00 5.89750498e+00 3.04832617e+00
2.83399619e-13 6.67744231e-14 4.44081103e-18 0.00000000e+00]
Condition number: 2.585991389306951e+21
||A p||_2: 0.0
Collinear case:
Design matrix shape: (40, 12)
Rank(A): 5 Nullity: 7
Singular values:
[6.82381053e+03 1.64481202e+03 6.44158346e+00 1.79053429e+00
2.15230664e-01 5.35073516e-14 1.87518157e-14 6.19959988e-16
1.00898999e-30 2.58571885e-31 0.00000000e+00 0.00000000e+00]
Condition number: inf
||A p||_2: 0.0
Too few points (5 world points -> 10 rows):
Design matrix shape: (10, 12)
Rank(A): 10 Nullity: 2
Singular values:
[3.74629383e+03 2.84267530e+03 9.91669999e+02 4.98023031e+02
3.95783162e+00 2.75987750e+00 1.17113458e+00 7.15905038e-01
2.96993304e-01 1.62792143e-01]
Condition number: 12614.068343471487
||A p||_2: 2.737838346337217e-13
--- Ill-Conditioned Demo ---
3D very clos:e
Design matrix shape: (80, 12)
Rank(A): 11 Nullity: 1
Singular values:
[5.10011841e+03 3.34694757e+01 2.62754421e+01 2.27855094e+01
6.32455891e+00 4.16352928e-02 3.30978595e-02 2.85353217e-02
7.02568928e-05 5.68713023e-05 4.92394028e-05 1.26925102e-15]
Condition number: 103577990.85941572
||A p||_2: 1.635441307864732e-14
3D very far apart
Design matrix shape: (80, 12)
Rank(A): 11 Nullity: 1
Singular values:
[2.63173530e+08 2.64640628e+07 5.51593155e+06 4.85409881e+04
4.23719991e+03 4.13017609e+03 3.24558117e+03 3.03838970e+03
2.75931754e+03 1.66415171e+01 5.90823329e+00 3.53065082e-12]
Condition number: 44543523.8074249
||A p||_2: 6.171896251171249e-09
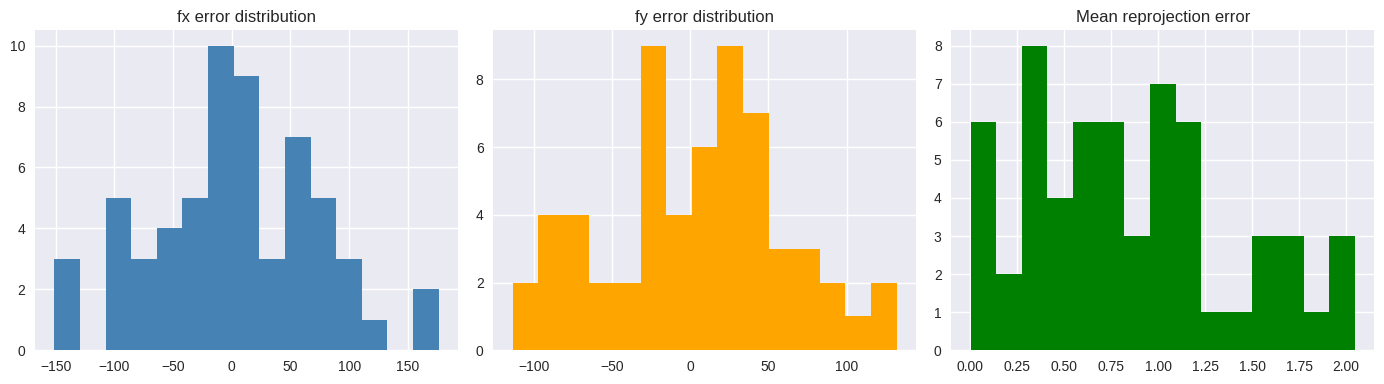
5.3.8. Noise Sensitivity and Monte Carlo Simulation#
We add Gaussian pixel noise and observe:
Intrinsic parameter errors
Reprojection errors
Distributions across trials
This quantifies robustness.
def add_noise(x, sigma=1.0):
rng = np.random.default_rng()
return x + rng.normal(0, sigma, x.shape)
def monte_carlo(n_trials=100, noise_sigma=1.0):
fx_errors, fy_errors, mean_reproj = [], [], []
for _ in range(n_trials):
x_noisy = add_noise(x_gt, noise_sigma)
A_n = build_dlt_matrix(X_world, x_noisy)
p_raw, _ = solve_dlt(A_n, analyze=False), _
P_tmp = vector_to_camera(p_raw)
K_tmp, R_tmp, t_tmp = decompose_camera_matrix(P_tmp)
stats_tmp, _ = reprojection_error(P_tmp, X_world, x_noisy)
fx_errors.append(K_tmp[0, 0] - K_gt[0, 0])
fy_errors.append(K_tmp[1, 1] - K_gt[1, 1])
mean_reproj.append(stats_tmp["mean"])
return np.array(fx_errors), np.array(fy_errors), np.array(mean_reproj)
fx_errs, fy_errs, reproj_means = monte_carlo(n_trials=60, noise_sigma=2.5)
plt.figure(figsize=(14, 4))
plt.subplot(1, 3, 1)
plt.hist(fx_errs, bins=15, color="steelblue")
plt.title("fx error distribution")
plt.subplot(1, 3, 2)
plt.hist(fy_errs, bins=15, color="orange")
plt.title("fy error distribution")
plt.subplot(1, 3, 3)
plt.hist(reproj_means, bins=15, color="green")
plt.title("Mean reprojection error")
plt.tight_layout()
plt.show()
print("fx error mean/std:", np.mean(fx_errs), np.std(fx_errs))
print("fy error mean/std:", np.mean(fy_errs), np.std(fy_errs))
print("Mean reprojection error mean/std:", np.mean(reproj_means), np.std(reproj_means))
fx error mean/std: 4.015350815073146 70.51873592945384
fy error mean/std: 4.170379344270911 57.39454663339191
Mean reprojection error mean/std: 0.8410928253290628 0.5435490517522839
